RADseq Genomics in R
computer setup
2025-09-02
Source:vignettes/rad_genomics_computer_setup.Rmd
rad_genomics_computer_setup.RmdTime to allow: ETA ~30-45 min for experience users, 1h-2h for novices
This vignette purposes is to show users how to setup their computers for RADseq genomic analysis inside R. It targets users of my packages or workshops.
The vignette as an Installation problems section, browse
through it if your experiencing installation issues.
The vignette is also usefull if your:
- conducting imputations
- using simulations with
grurand it’s dependencies or simulations withstrataG. - specific installation instructions for:
rmetasimXGBoostLightGBMrandomForestSRCrangermissRangerpcaMethods- how to have OpenMP enabled specific packages. R
packages that requires to run in parallel for the imputations
(e.g.
fstcore,fst,data.table).
I’m currently merging information also found in this tutorial
macOS
Warning: package management software Homebrew and/or MacPorts :
My experience with these packages is that at some point they will be unreliable with genomic software installation. It might do the trick for some software, but eventually you will lose a lot of time trying to figure out what’s your problem.
In this vignette I present brewless options only.
Update your OS
- the current version is macOS Sequoia 15.1
- how to update
Get administrator and root user access
Make sure you have administrator and root user access to your computer how.
Apple’s Command Line Tools
Make sure it’s installed…if not, follow instructions. Although the prompt message may be a bit confusing, just click install.
Notes:
- If command line tools is already installed you’ll get this message
xcode-select: error: command line tools are already installed, use "Software Update" to install updates - to test if you have a compiler, you can also type:
makein the terminal, if it’s installed you will have something similar to this*** No targets specified and no makefile found. Stop.
Terminology:
- Reading: to understand the basic steps to install software with the terminal (configure, make and sudo make install steps)
-
./configure: configure everything before installation -
make: connect libraries and the source before make install (doc) -
make install: use to build (compile) source code to create binary files and install the application on our system as configured using./configure, usually in/usr/local/bin.
Compiler
Apple can’t ship GNU Compiler Collection (GCC) with OpenMP enabled, similar story with Clang the other compiler used in macOS. Consequently, both need to be updated manually if you want to run software and packages that uses parallel computing (stacks, data.table, etc).
The command below fro both GCC and Clang will:
- change directory to your downloads
- download the compiler’s binaries
- uncompress and move folders (
bin,include,lib,libexecandshare) of the compiler on your computer/usr/localdirectory. - remove the downloaded file
- check the version of gcc
Update GCC
Choose the binary version number based on your OS and change the version accordingly link.
<<<<<<< HEAD Sonoma gcc-14.1-bin.tar.gz ======= Sonoma gcc-14.1-m1-bin.tar.gz >>>>>>> 625193a82a4a404c7c069f479c704172b2f74f16
#In Terminal
cd Downloads
<<<<<<< HEAD
sudo tar -zxvf gcc-14.1-bin.tar.gz -C /usr/local --strip-components=2Update Clang
We want clang compiler with OpenMP enabled. The latest version is 19.1.3.
PATH
The shell start up script and PATH to programs**
To make things a little easier to talk to your computer, each time you open the Terminal a shell start up scripts tells your computer where to look for programs. The path for your programs can be modified in your shell start up script. When your computer is searching for programs, it looks into these path:
The output should look like this:
/usr/bin:/bin:/usr/sbin:/sbin:/usr/local/bin. But
sometimes, it will also say: No such file or directory (no
worries, see below).
Use the pwd command to know exactly where you are!
The name of the shell startup file differs across platforms. Depending on OS it is called ~/.bash_profile and sometimes ~/.profile. Filename beginning with a dot “.” are reserved for the system and are invisible in the mac Finder.
Find your shell start up script with the following command:
If this returns nothing (blank), you don’t have a shell start up script. Create one with this command
To modify, you can use BBEdit to open or make and modify hidden items (using the option Show hidden items on the open file screen). Look for the free version. With Linux, use Vi!
Copy/paste the line below in your .bash_profile
file:
After modifying your shell start up script always run the command
source ~/.bash_profile to reload it.
Useful tools
Below are useful but not essential software you will like to have on your mac.
BBEdit
This is TextWrangler replacement and is a free text editor that will
help you save time. Once installed, go in the
Apple Menu bar -> BBEdit -> Install Command Line
pip
This is a Python installer tool that I highly recommend. To install or upgrade pip, securely download get-pip.py.
Run this from you Terminal:
If you get command not found you might not have python installed.
Linux
Make sure you have GCC and CLANG with OpenMP enabled. Several flavors available, check for the proper link
Install R
To install R v4.4.2 “Pile of Leaves” released on 2024-10-31 download the installer and follow the instructions
To remove R completely from macOS
Rstudio
To download RStudio, check this link and download the installer for your OS.
Installing Libraries
Below is how I setup most of my computers after a clean macOS install. 1. Start with devtools and tidyverse
if (!require("devtools")) install.packages("devtools") # to install
install.packages("tidyverse")- The gsl package is required by numerous packages and can cause some headaches… Don’t install it from source.
install.packages("gsl")If the console print this:
Do you want to install from sources the package which needs compilation? (Yes/no/cancel).
Always aswer no unless of course you know what you are
doing.
- Than, I install grur and assigner my packges with the higher number of dependencies, this way it’s installing automatically 90% of the packages I need (including radiator).
devtools::install_github("thierrygosselin/grur")
devtools::install_github("thierrygosselin/assigner")Makevars file
For some packages you might have to compile from source and the use
of different compiler is sometimes very useful. You need to tell R how
to use the compilers. This might change from one package to another.
Nothing is simple, you know this by now… All this is done through R’s
Makevars file located in
~/.R/Makevars.
To modify or create the file, the fastest way is to use the package
usethis (it’s installed
automatically with devtools):
usethis::edit_r_makevars()Makevars content required:
CC=/usr/local/bin/gcc
CXX=/usr/local/bin/g++
FC=/usr/local/bin/gfortran
F77=/usr/local/bin/gfortran
PKG_LIBS = -fopenmp -lgomp
PKG_CFLAGS= -O3 -Wall -pipe -pedantic -std=gnu99 -fopenmp
PKG_CXXFLAGS=-fopenmp -std=c++11
CFLAGS= -O3 -Wall -pipe -pedantic -std=gnu99 -fopenmp
SHLIB_OPENMP_CFLAGS = -fopenmp
SHLIB_OPENMP_CXXFLAGS = -fopenmp
SHLIB_OPENMP_FCFLAGS = -fopenmp
SHLIB_OPENMP_FFLAGS = -fopenmp
# change the nex line according to your computer compiler version (use gcc -v in terminal):
FLIBS=-L/usr/local/lib/gcc/x86_64-apple-darwin19/9.2.0/finclude
CFLAGS=-mtune=native -g -O2 -Wall -pedantic -Wconversion
CXXFLAGS=-mtune=native -g -O2 -Wall -pedantic -WconversionErrors and installation problems
Sometimes you’ll get warnings while installing dependencies required for x package.
#Warning: cannot remove prior installation of package ‘stringi’To solve this problem, delete manually the problematic package in the
installation folder (on mac:
/Library/Frameworks/R.framework/Resources/library) or in
the Terminal:
sudo rm -R /Library/Frameworks/R.framework/Resources/library/package_name
# Changing 'package_name' to the problematic package.
# Reinstall the package.Using the latest version of R, RStudio and packages is recommended. If your heart start pounding just at the thought of having to install a new R version, you should have a look at packrat.
Look at the output in R console when you get an error message. If it’s related to one’s of the packages dependencies, try installing it separately before attempting to reinstall the problematic package.
Error: macOS Big Sur update
So far, I’ve only experience 1 problem after upgrading to Big Sur, and it’s linked to adegenet dependency on sf package (solution).
Error: checking whether the C++ compiler works… no
When rtying to compile a software if you get this error:
checking whether the C++ compiler works... no or
configure: error: C++ compiler cannot create executables
Try installing Xcode from the App Store.
Error: “https” not supported
If you get something like:
https not supported or disabled in libcurl, install or
re-install:
Required if GCC compiler is used (TLS backend is then used). Not required if clang is used (securetransport backend is used).
#In browser
https://www.openssl.org/source/openssl-1.1.1.tar.gz
#In Terminal
cd ~/Downloads
curl -L https://www.openssl.org/source/openssl-1.1.1.tar.gz | tar xf -
cd openssl-1.1.1
./config
make -j12 #change with your number of CPU
make test #long
sudo make install
cd ..
sudo rm -R openssl*Check for the latest release of curl
#Copy/paste in your browser
https://curl.haxx.se/download/curl-7.85.0.tar.gz
# Terminal
cd ~/Downloads
tar -zxvf curl-7.85.0.tar.gz
cd curl-7.85.0for more curl* option type curl -h*
The next step depends on the compiler used
With gcc:
Note: with macOS 11.0.1 this give me an error
configure: error: OpenSSL libs and/or directories were not found where specified!
If this is the case, use clang:
Linux related problem
If you have an install problem, the problem might be very
computer-specific. e.g. if the problem is related to
strataG, copula and/or gsl, try
installing libgsl0-dev in the Terminal
(very easy now with the latest RStudio release!):
Error: vector memory exhausted
For errors that highlight problems with vectors and memory similar
to: vector memory exhausted (limit reached). In R, verify
that you have a file called ~/.Renviron:
file.exists("~/.Renviron")If you don’t have the file:
Add this to your .Renviron file located in
~/.Renviron:
You can also use a text editor that allows you to see hidden files
(files starting with a . dot).
Error during package installation
- Problem with
string.h,math.hor any other.h - Problem with
vroom - Problem with
rmetasim
An example during vroom installation:
clang++ -std=gnu++11 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I"/Library/Frameworks/R.framework/Versions/3.5/Resources/library/Rcpp/include" -I"/Library/Frameworks/R.framework/Versions/3.5/Resources/library/progress/include" -I/usr/local/include -Imio/include -DWIN32_LEAN_AND_MEAN -Ispdlog/include -fPIC -Wall -g -O2 -c gen.cc -o gen.o
clang++ -std=gnu++11 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I"/Library/Frameworks/R.framework/Versions/3.5/Resources/library/Rcpp/include" -I"/Library/Frameworks/R.framework/Versions/3.5/Resources/library/progress/include" -I/usr/local/include -Imio/include -DWIN32_LEAN_AND_MEAN -Ispdlog/include -fPIC -Wall -g -O2 -c index_collection.cc -o index_collection.o
/usr/local/bin/clang -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I"/Library/Frameworks/R.framework/Versions/3.5/Resources/library/Rcpp/include" -I"/Library/Frameworks/R.framework/Versions/3.5/Resources/library/progress/include" -I/usr/local/include -fPIC -I/usr/local/include -c localtime.c -o localtime.o
localtime.c:42:10: fatal error: 'string.h' file not found
#include <string.h>
^~~~~~~~~~With macOS : your Makevars needs
additional lines to make it work configuration.
Error .setMaxGlobalSize
Its a problem with a previous version of pbmcapply and
it’s interaction with future.
Solution:
- restart your computer
- with a clean desk and session: open RStudio and update your packages
- Restart R (
RStudio > Session > Restart R) - Re-install my github package generating the error
Note: if your heart start pounding just at the thought of having to update everything on your computer you should definitely have a look at packrat: it’s very easy to use.
Error .DynamicClusterCall
If you have a PC and you’re getting this error or closely related error:
# Error in .DynamicClusterCall(cl, length(cl), .fun = function(.proc_idx, :
# One of the nodes produced an error: Can not open file 'FILE PATH'. The process cannot access the file because it # is being used by another process.Solution: Use parallel.core = 1 in the
function generating the error.
Corrupt packages
The error below usually happens when several packages are updated:
# Error in get0(oNam, envir = ns) :
# lazy-load database '/Library/Frameworks/R.framework/Versions/3.5/Resources/library/callr/R/callr.rdb' is corrupt
# In addition: Warning message:
# In get0(oNam, envir = ns) : internal error -3 in R_decompress1Solution:
- restart R
- re-install the package that generated this error
Error: dyn.load
I see 2 problems and separate solutions. When the error is related to adegenet, strataG, assigner or packages that depends on sf package, see the solution below.
When the error is similar to:
Error in dyn.load(file, DLLpath = DLLpath, ...) :
unable to load shared object '/home/rstudio/R/x86_64-pc-linux-gnu-library/3.6/units/libs/units.so':
libudunits2.so.0: cannot open shared object file: No such file or directory
Calls: <Anonymous> ... asNamespace -> loadNamespace -> library.dynam -> dyn.load
Execution haltedSolution:
- In R, check the output of this command that look for the environment variables:
Sys.getenv("LD_LIBRARY_PATH")
# [1]""Note that if the output is not empty, like in the example above, write down the output.
- Set the new environment
variables by adding
/usr/local/lib/to the output above:
When it’s empty:
# in R:
Sys.setenv(LD_LIBRARY_PATH="/usr/local/lib/")
# For Linux you could use: /usr/local/lib/:/usr/lib64 When it’s not, add at the end, separated by :
Sys.setenv(LD_LIBRARY_PATH="/usr/local/lib64/R/lib:/lib:/usr/local/lib64:/usr/lib/jvm/java-1.8.0-openjdk-1.8.0.222.b10-0.amzn2.0.1.x86_64/jre/lib/amd64/server:/usr/local/lib/:/usr/lib64")Long-term solution:
Instead of using Sys.setenv each time you have a similar
problem, you could add the environment
variables LD_LIBRARY_PATH to your
.Renviron file. This is discussed in
another problem above.
Error related to sf
and dyn.load
Sometimes the problem related to dyn.load problems originates from specific packages. So far, for me, they are always linked to sf package. The problem for macOS is usually when updating os, e.g. from Catalina -> Big Sur. The solution is similar for Linux when you try to install adegenet.
There are solutions using Homebrew, but I’m not a fan for the shortcuts.
brewless version to execute in the terminal:
cd ~/Downloads
curl -L https://downloads.unidata.ucar.edu/udunits/2.2.28/udunits-2.2.28.tar.gz | tar xf -
cd udunits*
./configure && make -j14 && sudo make install
cd ..
sudo rm -R udunits*cd ~/Downloads
curl -L https://www.sqlite.org/2025/sqlite-autoconf-3490100.tar.gz | tar xf -
cd sqlite-autoconf*
./configure && make -j14 && sudo make install
cd ..
sudo rm -R sqlite-autoconf*cd ~/Downloads
curl -L https://download.osgeo.org/libtiff/tiff-4.7.0.tar.gz | tar xf -
cd tiff*
#CC=/usr/local/bin/clang
#CXX=/usr/local/bin/clang++
./configure
#./configure --prefix=/usr/local/ --host=aarch64 --libdir=/usr/local/lib
make -j14
# not working on M4 macs
sudo make install
cd ..
sudo rm -R tiff*cd ~/Downloads
curl -L https://download.osgeo.org/proj/proj-9.5.1.tar.gz | tar xf -
cd proj*
mkdir build
cd build
cmake ..
cmake --build .
sudo cmake --build . --target install
cd ~/Downloads
sudo rm -R proj*
# with prior releases, when configure was necessary
# ./configure --libdir=/usr/local/lib && make -j56 && sudo make install export LDFLAGS="-L/usr/local/lib"
export CPPFLAGS="-I/usr/local/include"
cd ~/Downloads
curl -L https://github.com/OSGeo/gdal/releases/download/v3.5.3/gdal-3.5.3.tar.gz | tar xf -
cd gdal*
mkdir build
cd build
cmake ..
cmake --build .
sudo cmake --build . --target install
cd ~/Downloads
sudo rm -R gdal*
# previous install command required
#./configure --prefix=/usr/local --libdir=/usr/local/lib --with-proj=/usr/local && make -j56 && sudo make install #time for coffee...cd ~/Downloads
git clone https://git.osgeo.org/gitea/geos/geos.git
cd geos*
mkdir build
cd build
cmake ..
cmake --build .
sudo cmake --build . --target install
cd ~/Downloads
sudo rm -R geos*
# previous install command required
#./autogen.sh
#./configure --libdir=/usr/local/lib && make -j56 && sudo make installBack in R/RStudio
install.packages("rgeos", repos="http://R-Forge.R-project.org", type="source")
install.packages("rgdal", repos="http://R-Forge.R-project.org", type="source")
devtools::install_github("r-spatial/sf", configure.args = "--with-proj-lib=/usr/local")
install.packages("adegenet")Error: HTTP status was 404 Not Found
This error is sometimes poping after a new R upgrade. Try installing the problematic package differently.
Instead of:
BiocManager::install("SeqArray")Try:
remotes::install_local(path = "SeqArray_latest.tar.gz")Error: C stacks usage
So far, I haven’t found the cure to this computer-specific problem.
Potential solutions:
- Make sure you have enough RAM on your computer for the dataset you want to analyze (16GB is the bare minimum these days for small-medium RADseq dataset).
- Closing all windows/app running in the background somtimes help.
- Updating to the latest version of R, RStudio and libraries.
- Not a fan of upgrading to latest version, use Packrat.
- Follow the guidelines to prepare your computer in this vignette, from the start…
Specific librairies installation
fst & fstcore &
data.table
Better to install and compile them from source to enable OpenMP. Install in the terminal zstd and lz4:
cd ~/Downloads
curl -L https://github.com/lz4/lz4/archive/refs/tags/v1.10.0.tar.gz | tar xf -
cd lz4*
make
sudo make install
cd ..
sudo rm -R lz4*cd ~/Downloads
curl -L https://github.com/facebook/zstd/archive/refs/tags/v1.5.6.tar.gz | tar xf -
cd zstd*
make
sudo make install
cd ..
sudo rm -R zstd*fstcore, fst and data.table
requires these R Makevars specifications. If you have other
lines, comment (#) before saving and installing from
source:
usethis::edit_r_makevars()#fstcore fst data.table
CC=/usr/local/bin/gcc -fopenmp
CXX=/usr/local/bin/g++ -fopenmp
CXX11=/usr/local/bin/g++ -fopenmp
CXX14=/usr/local/bin/g++ -fopenmp
CXX17=/usr/local/bin/g++ -fopenmp
CXX1X=/usr/local/bin/g++ -fopenmp
CXX98=/usr/local/bin/g++ -fopenmp
install.packages("fstcore", type = "source")
install.packages("fst", type = "source")
install.packages("data.table", type = "source")
rmetasim
Download
Download the latest github release of Allan Strand’s rmetasim
Modify the number of loci: MAXLOCI
If you want to use more loci during your simulations (default is
10001), you need to modify rmetasim before compiling. With a text
editor, modify the const.h file in the src
folder: rmetasim-master/src/const.h. Navigate to
lane 33 and change the integer to the desired maximum
number of loci. Or do this in the Terminal:
Makevars configuration
rmetasim requires these Makevars (~/.R/Makevars
file) specifications. If you have other lines, comment (#) before
compiling rmetasim:
usethis::edit_r_makevars()macOS
This latest os requires extra lines:
CC=/usr/local/bin/clang
CXX=/usr/local/bin/clang++
CXX1X=/usr/local/bin/clang++
FLIBS=-L/usr/local/lib
LDFLAGS=-L/usr/local/lib
SHLIB_OPENMP_CFLAGS= -fopenmp
SHLIB_OPENMP_FCFLAGS= -fopenmp
SHLIB_OPENMP_FFLAGS= -fopenmp
SHLIB_OPENMP_CXXFLAGS= -fopenmp
CFLAGS+=-isysroot /Library/Developer/CommandLineTools/SDKs/MacOSX.sdk
CCFLAGS+=-isysroot /Library/Developer/CommandLineTools/SDKs/MacOSX.sdk
CXXFLAGS+=-isysroot /Library/Developer/CommandLineTools/SDKs/MacOSX.sdk
CPPFLAGS+=-isysroot /Library/Developer/CommandLineTools/SDKs/MacOSX.sdk
XGBoost
Error?
If you’re getting this error:
"https" not supported or disabled in libcurl, extra steps
are required, check the installation
problems section to install OpenSSL and
curl and enabling https with
--with-ssl option.
Makevars configuration
XGBoost requires these Makevars specifications. If you
have other lines, comment (#) before compiling:
usethis::edit_r_makevars()CC=/usr/local/bin/gcc
CXX=/usr/local/bin/g++
CXX11=/usr/local/bin/g++
CXX14=/usr/local/bin/g++
CXX17=/usr/local/bin/g++
SHLIB_OPENMP_CFLAGS= -fopenmp
SHLIB_OPENMP_FCFLAGS= -fopenmp
SHLIB_OPENMP_FFLAGS= -fopenmp
SHLIB_OPENMP_CXXFLAGS= -fopenmp
CFLAGS=-g -O3 -Wall -pedantic -std=gnu99 -mtune=native -pipe
CXXFLAGS=-g -O3 -Wall -pedantic -std=c++11 -mtune=native -pipe
LDFLAGS=-L/usr/local/lib -Wl,-rpath,/usr/local/lib
CPPFLAGS=-I/usr/local/include -I/usr/local/includeTest R-package
You should see a time difference between both runs
require(xgboost)
x <- matrix(rnorm(100*10000), 10000, 100)
y <- x %*% rnorm(100) + rnorm(1000)
system.time({bst = xgboost(data = x, label = y, nthread = 1, nround = 100, verbose = FALSE)})
system.time({bst = xgboost(data = x, label = y, nthread = 4, nround = 100, verbose = FALSE)})
LightGBM
LightGBM requires an OpenMP-enabled compiler. Currently,
it doesn’t work well with clang, so make sure you have updated your GCC
compiler (instructions above). Additionally, LightGBM
requires CMake
Download and install CMake
#In browser or using curl in Terminal
https://github.com/Kitware/CMake/releases/download/v4.0.0/cmake-4.0.0-macos-universal.dmg
# double-click on the disk image and follow instructionsTo add CMake to the PATH:
PATH="/Applications/CMake.app/Contents/bin":"$PATH"
# Or, to install symlinks to '/usr/local/bin', run:
sudo "/Applications/CMake.app/Contents/bin/cmake-gui" --install
# Or, to install symlinks to another directory, run:
sudo "/Applications/CMake.app/Contents/bin/cmake-gui" --install=/path/to/bin
#Then, run the following commands to install LightGBM:
randomForestSRC
randomForestSRC requires the GCC OpenMP-enabled compiler to run in parallel. See instructions above if not already done.
Makevars configuration
Check that the lines below are not commented in your
~/.R/Makevars file:
usethis::edit_r_makevars()CC=/usr/local/bin/gcc
CXX=/usr/local/bin/g++
CFLAGS=-g -O3 -Wall -pedantic -std=gnu99 -mtune=native -pipe
CXXFLAGS=-g -O3 -Wall -pedantic -std=c++11 -mtune=native -pipe
PKG_CFLAGS= -O3 -Wall -pipe -pedantic -std=gnu99 -fopenmp
PKG_CXXFLAGS=-fopenmp -std=c++11
FC=/usr/local/bin/gfortran
F77=/usr/local/bin/gfortran
LDFLAGS=-L/usr/local/lib
PKG_LIBS = "-liconv"Download and install
From the Terminal run these steps to download and compile randomForestSRC:
cd ~/Downloads
curl -L https://cran.r-project.org/src/contrib/randomForestSRC_2.9.3.tar.gz | tar xf -
cd randomForestSRCMake sure you have autoconf installed:
Should output: autoconf: error: no input file, if not,
install following the steps here.
# in Terminal
cd ~/Downloads/randomForestSRC
autoconf
cd ~/Downloads
R CMD INSTALL --preclean --clean randomForestSRCYou want to make sure that this line is printed during execution of
the previous command:
checking whether OpenMP will work in a package... yes or
checking for /usr/local/bin/gcc option to support OpenMP... -fopenmp
fastsimcoal2
To install fastsimcoal2
v.2.6.0.3, to use in grur::simulate_rad:
COLONY
To install COLONY 30/07/2018, V2.0.6.5:
MacOS
The old openmpi version (openmpi-1.6.5) is required, saddly.
cd ~/Downloads
curl -L https://download.open-mpi.org/release/open-mpi/v1.6/openmpi-1.6.5.tar.gz | tar xf -
cd openmpi-1.6.5
export TMPDIR=/tmp
./configure F77=gfortran #--prefix=/usr/local -openmp # no longer work for some reason
make -j 12
sudo make install
sudo rm -R ~/Downloads/openmpi*To download COLONY, follow instructions on Jinliang Wang ZSL website. The
file you need to uncompress is named:
colony2.mac_.20180730.zip.
Linux
To download COLONY, follow instructions on Jinliang Wang ZSL website. The
file you need to uncompress is named:
colony2.linux_.20180730.zip.
Several options are available depending on the compiler you have installed.
Useful
Update Github
macOS comes with Github, a Version
Control System (VCS), pre-installed. However, the install is in
/usr/bin/git which can make it difficult for beginners to
update. To change this, run these commands:
- Install GNU Autoconf
cd ~/Downloads
curl -L http://ftp.gnu.org/gnu/autoconf/autoconf-latest.tar.gz | tar xf -
cd autoconf-2.69
./configure
make
sudo make install
cd ..
sudo rm -R ~/Downloads/autoconf-*- Install Github
git --version # show current git version installed
which git # returns where is git on your computer
cd ~/Downloads
git clone https://github.com/git/git # install the latest Git
cd git
make configure
./configure
make -j12
sudo make install
cd ..
sudo rm -R ~/Downloads/git/ # remove git folder
source ~/.bash_profile # reload startup script
git --version # confirmed the version you just installed
which git # returns /usr/local/binmacOS Terminal from specific folder
In System Preferences choose
Keyboard -> Shortcuts. From the left panel, choose
Services. In the right panel, under Files and
Folders, choose New Terminal at Folder and/or New
Terminal Tab at Folder. Now you can right-click your track pad
or mouse on a folder and choose Services -> New Terminal at
Folder!
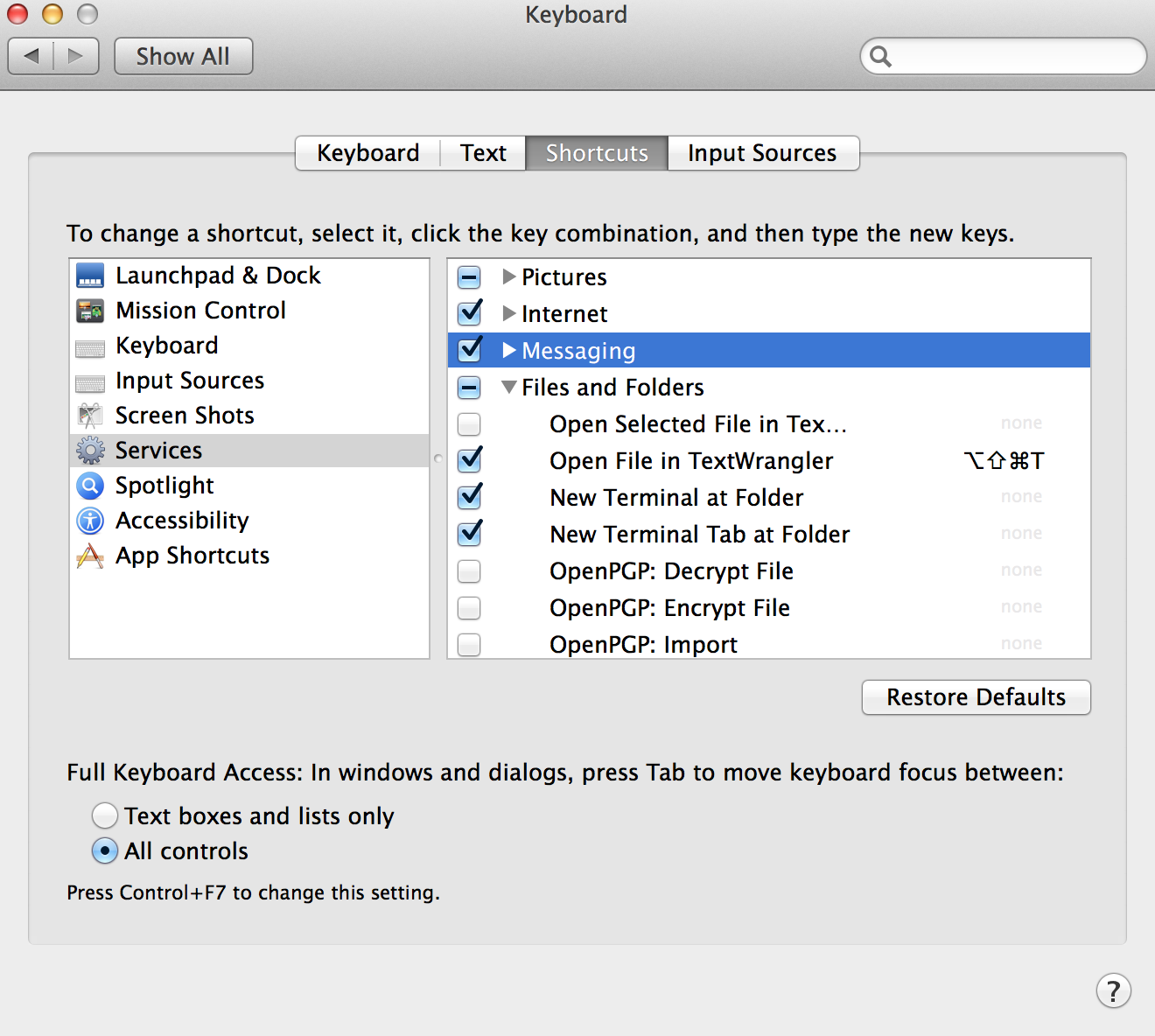
Shortcut to folder path
With macOS, open the Automator application.
File -> New (cmd-N)
Choose: Services
Left panel, choose: Library -> Utilities
Middle, choose: Copy to Clipboard and drag it to the right panel
Now you want to have: Service receives selected FILES OR FOLDERS in FINDER>
You should have something similar to the image below: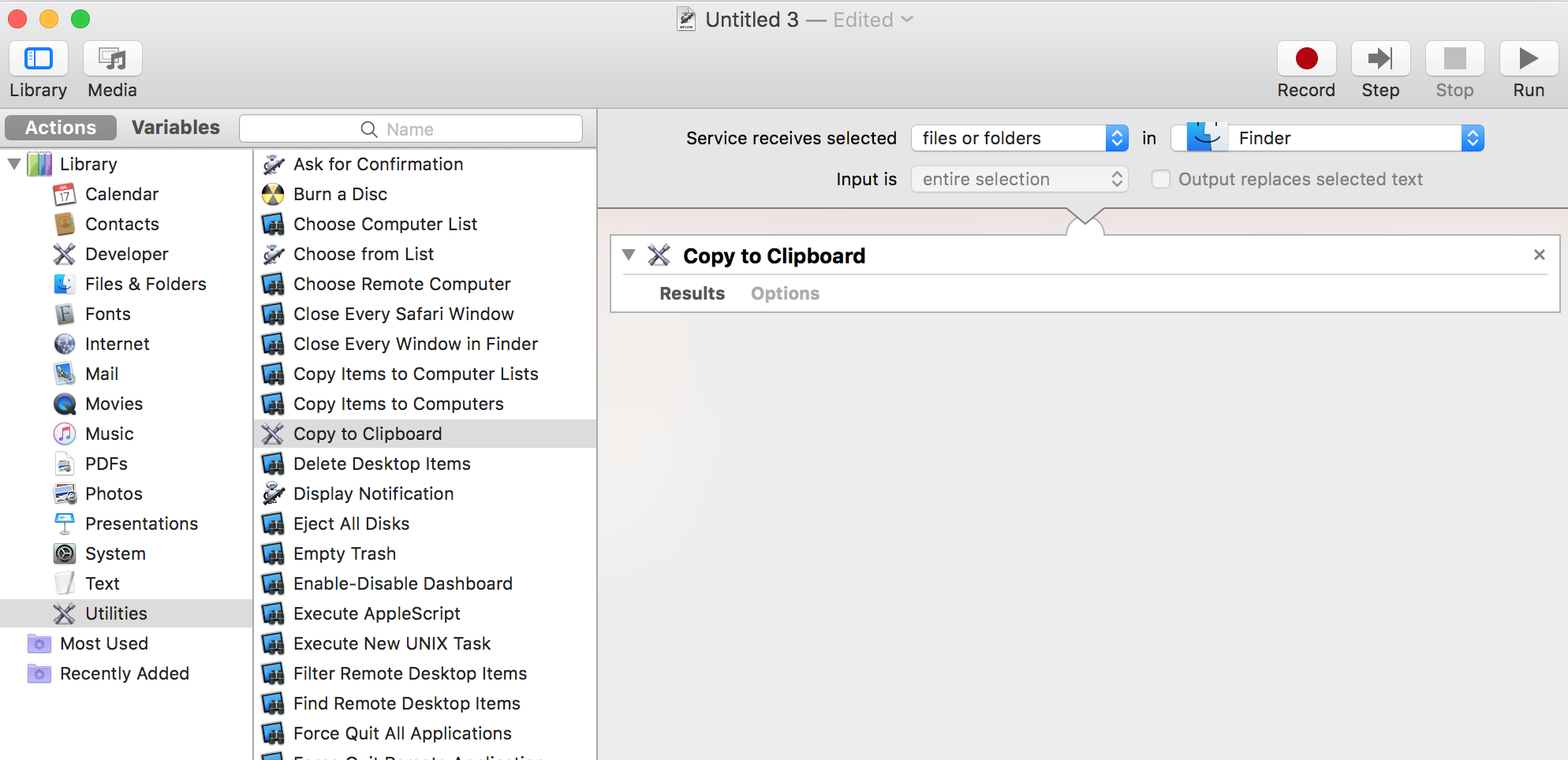
Go in the Finder, select a folder and right click on it you should see ‘copy path to clipboard’ at the bottom or in Services.