This is the development page of the stackr.
What’s the difference with running stacks directly in the terminal?
Besides running stacks within R, not much, tiny differences here and there that speed up my RADseq workflow:
- The philosophy of working by project with pre-organized folders.
- Some important steps are parallelized.
- You have more than 1 sequencing chip/lane ? This workflow will save you lots of time.
- Technical replicates, inside or across chip/lanes are managed uniquely.
- Noise reduction.
- Data normalization.
- nightmares because of a crashed computer/cluster/server? stackr manage stacks unique integer (previously called SQL IDs) throughout the pipeline. It’s integrated from the start, making it a breeze to just re-start your pipeline after a crash!
-
mismatch testing: de novo mismatch threshold series is integrated inside
run_ustacksand stackr will produce tables and figures automatically. - catalog: for bigger sampling size project, breaking down the catalog into several separate cstacks steps makes the pipeline more rigorous if your computer/cluster/server crash.
- logs generated by stacks are read and transferred in human-readable tables/tibbles. Detecting problems is easier.
- summary of different stacks modules: available automatically inside stackr pipeline, but also available for users who didn’t use stackr to run stacks.
- For me all this = increased reproducibly.
Who’s it for?
- It’s currently developed with my own projects in mind.
- To help collaborators to get the most out of stacks.
It’s not for R or stacks beginners. stacks related issues should be highlighted on stacks google group.
Installation
To try out the dev version of stackr, copy/paste the code below:
if (!require("devtools")) install.packages("devtools")
devtools::install_github("thierrygosselin/stackr")
library(stackr)Citation:
To get the citation, inside R:
citation("stackr")Web site with additional info: http://thierrygosselin.github.io/stackr/
Life cycle
stackr is maturing, but in order to make the package better, changes are inevitable. Argument names are very stable and follows stacks development closely.
- Philosophy, major changes and deprecated functions/arguments are documented in life cycle section of functions.
- The latest changes are documented in changelog, versions, new features and bug history
- issues and contributions
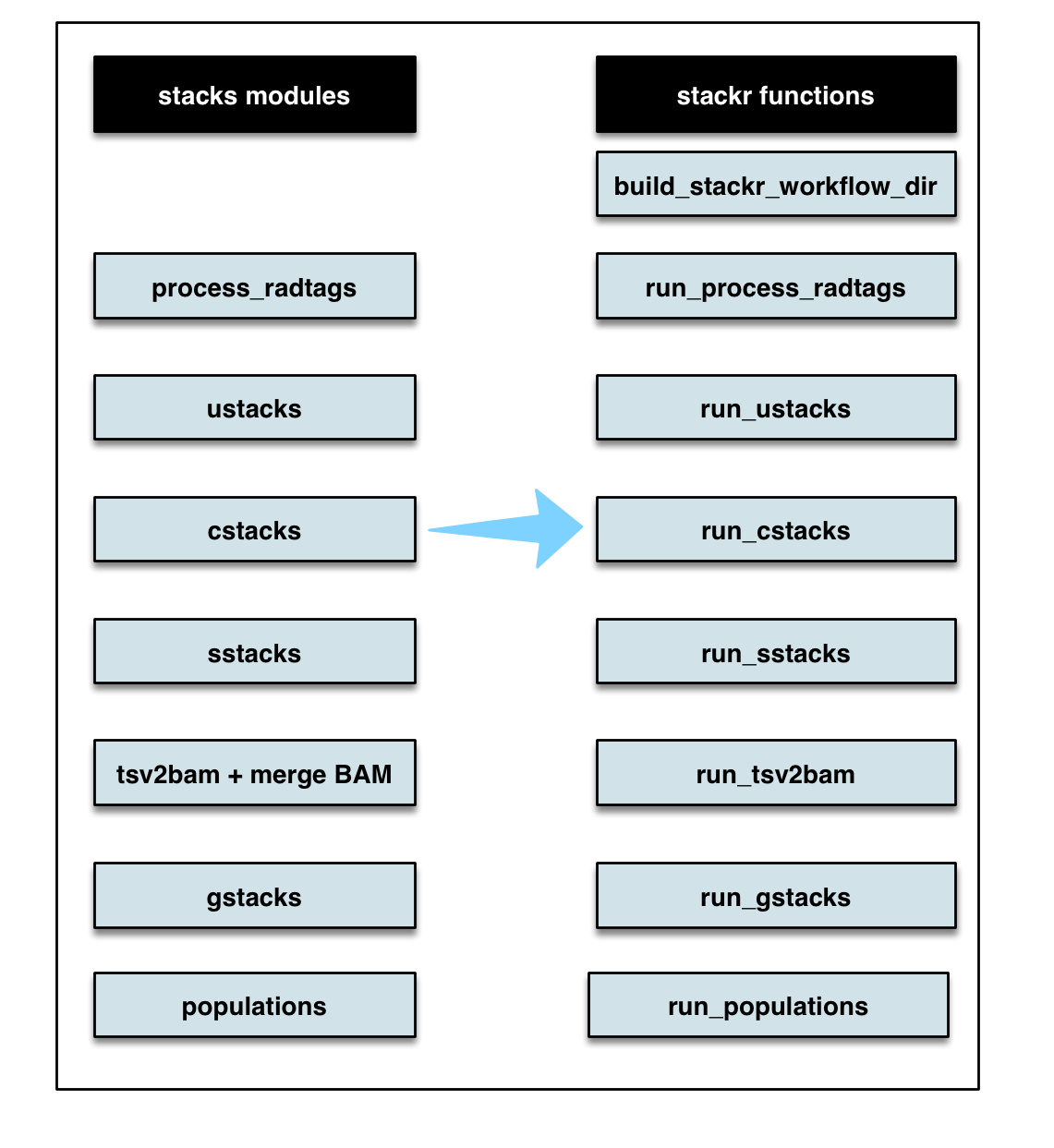
Stacks modules and RADseq typical workflow
stackr package provides wrapper functions to run STACKS process_radtags, ustacks, cstacks, sstacks, rxstacks and populations inside R.
Below, a flow chart showing the corresponding stacks modules and stackr corresponding functions.