stackr: an R package to run stacks pipeline
This is the development page of stackr.
stackr package provides wrapper functions to run STACKS (process_radtags, ustacks, cstacks, sstacks, tsv2bam, gstacks and populations) inside R.
Who is it for ?
- stackr is currently developed for my projects at CSIRO and to help colleagues and collaborators to get the most out of stacks.
- I make stackr available for others, but I don’t offer help, use it at your own risk.
- STACKS related issues should be highlighted in stacks google group.
- It’s not for R and/or stacks beginners. Train with STACKS in the Terminal first.
Overview of the differences from running stacks
- I work with thousands of samples, my STACKS experience is more reproducible and organized inside RStudio
- stackr requires you to think ahead by using a project info file that outlines the sample names and it’s metadata (think sampling sites, barcodes, sequencing lanes, etc).
- The outputs are organized by folders, it’s more tidy and it’s automatic. e.g. logs generated by STACKS are summarized in human-readable tables/tibbles, making problem detection easier.
- Most functions that runs STACKS can be restarted if the computer/cluster/server crash. Just restart the same function. stackr takes care of the rest.
Made it this far, here’s more details:
-
process_radtags ->
run_process_radtags(demultiplexing) : works better with multiple lanes and technical replicates inside or across chip/lanes are managed and accounted for. -
ustacks ->
run_ustacks(de novo assembly): manages STACKS unique integer (previously called SQL IDs) and can also do mismatch testing! Integrated insiderun_ustacksis a de novo mismatch threshold series function that generates tables and figures automatically to help you pick the appropriate thresholds. -
cstacks ->
run_cstacks(catalog)- With more than 1000 samples, cstacks is problematic: time consuming and vulnerable to computer crashes and other interruptions.
- stackr generates catalogs incrementally, this way the pipeline is more rigorous if your computer/cluster/server crash, because the previous catalog will already be saved and available to start over.
- Different catalogs with increasing sample number = better testing
Installation
To try out the dev version of stackr, copy/paste the code below:
if (!require("devtools")) install.packages("devtools")
devtools::install_github("thierrygosselin/stackr")
library(stackr)Stacks modules and RADseq typical workflow
stackr package provides wrapper functions to run STACKS process_radtags, ustacks, cstacks, sstacks, tsv2bam, gstacks and populations inside R.
Below, a flow chart showing the corresponding stacks modules and stackr corresponding functions. 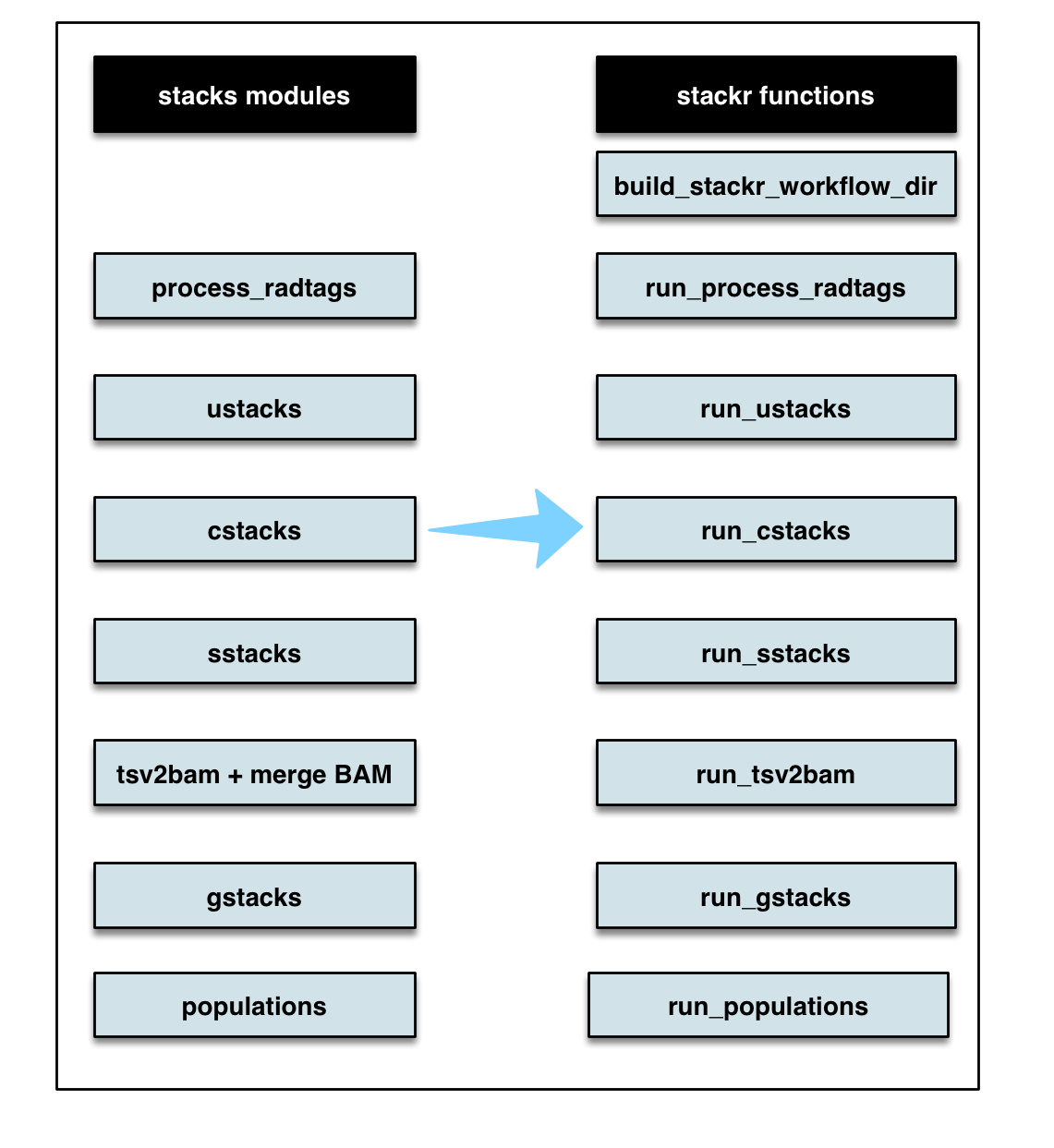
Vignette
- Get started section
- Web site with additional info: stackr
- Computer setup and troubleshooting
- Vignettes
Life cycle
stackr is quite mature, i’ve used it for almost 8 years with dozens of projects, but changes are inevitable.
- For reproducibility, if you use it, keep a copy safe.
- I follow STACKS development closely, when a new version comes out, stackr will likely change as well.
- Because of it’s intrinsic nature, stackr will not be compatible with previous STACKS version.
- Philosophy, major changes and deprecated functions/arguments are documented in life cycle section of functions.
- The latest changes are documented in changelog, versions, new features and bug history
- issues and contributions




